MultiDendrograms
A hierarchical clustering tool
Index
- Description
- Comparison with other applications
- References
- Download
- Requirements
- Installation
- Development
- Gallery
- History
- Authors
Description
MultiDendrograms is a simple yet powerful program to make the Hierarchical Clustering of real data, distributed under an Open Source license. Starting from a distances (or similarities) matrix, MultiDendrograms calculates its dendrogram using the most common Agglomerative Hierarchical Clustering algorithms, allows the tuning of many of the graphical representation parameters, and the results may be easily exported to file. A summary of characteristics:
- Multiplatform: developed in Java, runs in all operating systems (e.g. Windows, Linux and MacOS).
- Graphical user interface: data selection, hierarchical clustering options, dendrogram representation parameters, navigation across the dendrogram, deviation measures.
- Hierarchical Clustering algorithms: weighted and unweighted variable-group versions of Single Linkage, Complete Linkage, Arithmetic Linkage (UPGMA), Versatile Linkage, Centroid, Ward and Beta Flexible.
- Representation parameters: size, orientation, labels, axis, etc.
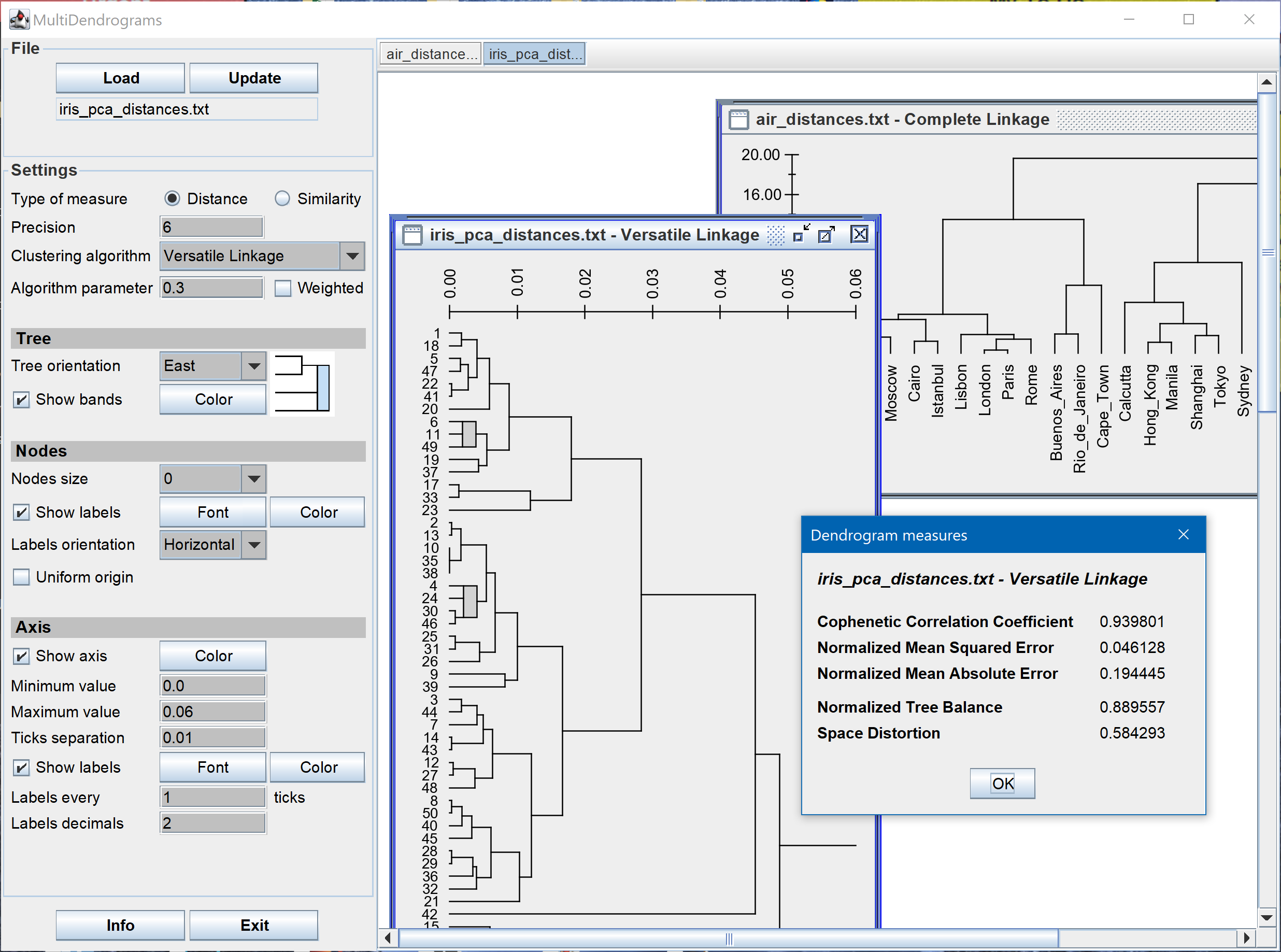
- Dendrogram measures: Tree Balance, Cophenetic Correlation Coefficient, Normalized Mean Squared Error, Normalized Mean Absolute Error and Space Distortion.
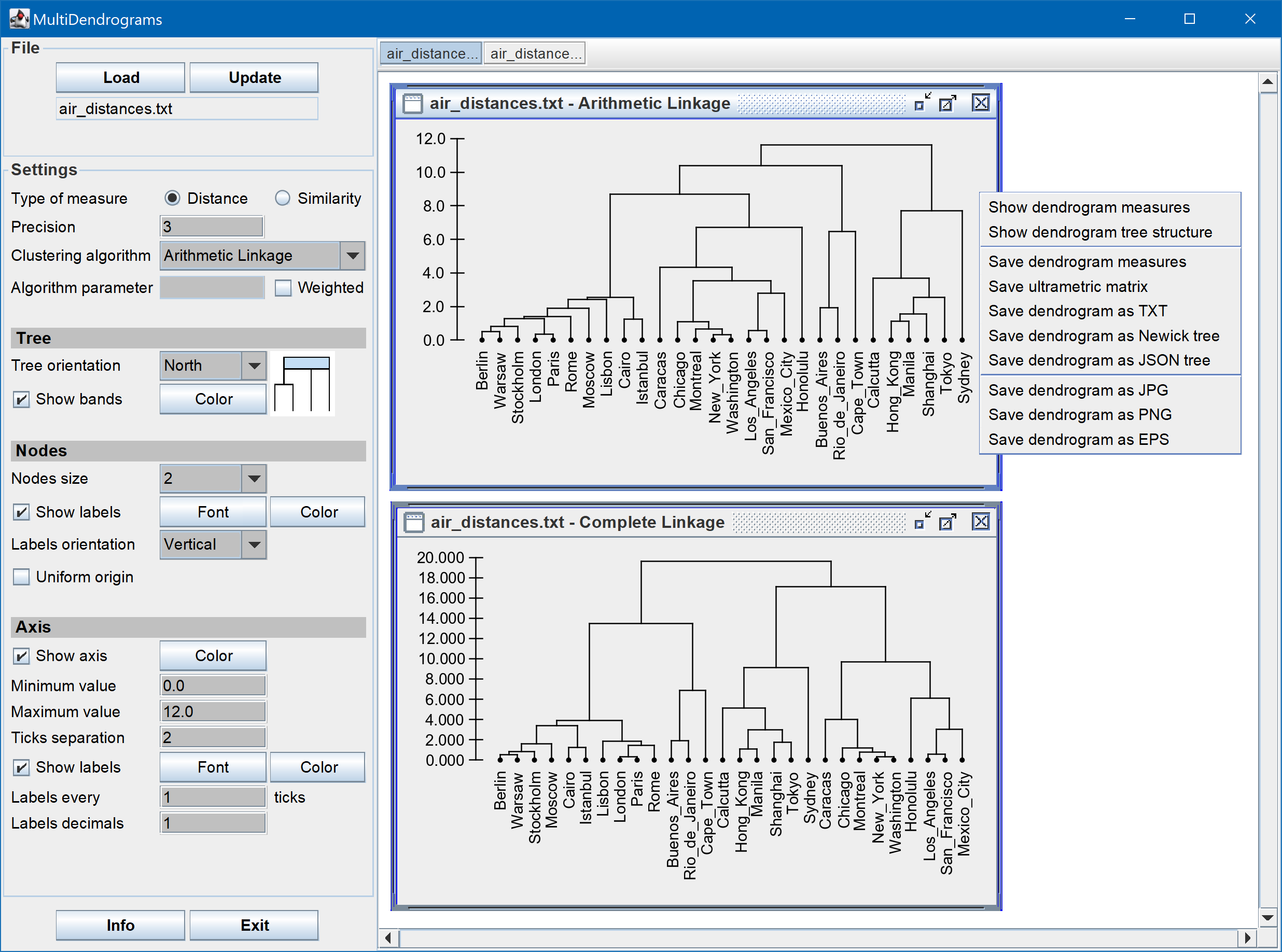
- Export: ultrametric matrix, dendrogram measures, dendrogram details in text, Newick and JSON tree formats.
- Plot: dendrogram image in JPG, PNG and EPS formats.
- Command-line: available direct calculation of hierarchical clustering from the command-line, without the need to use the graphical interface.
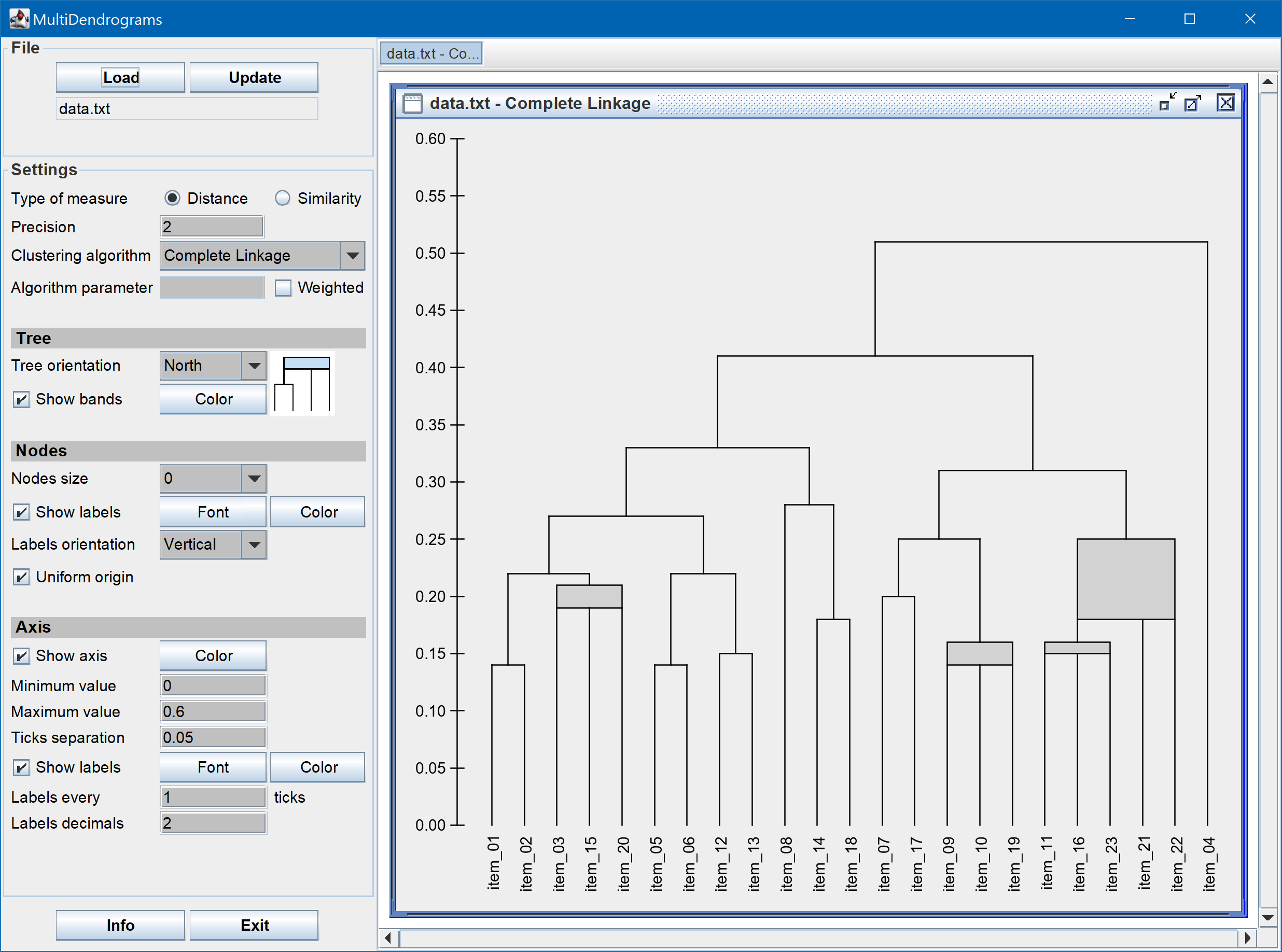
MultiDendrograms implements the variable-group algorithms in [1] to solve the non-uniqueness problem found in the standard pair-group algorithms and implementations. This problem arises when two or more minimum distances between different clusters are equal during the amalgamation process. The standard approach consists in choosing a pair, breaking the ties between distances, and proceeds in the same way until the final hierarchical classification is obtained. However, different clusterings are possible depending on the criterion used to break the ties (usually a pair is just chosen at random!), and the user is unaware of this problem.
The variable-group algorithms group more than two clusters at the same time when ties occur, given rise to a graphical representation called multidendrogram. Their main properties are:
- When there are no ties, the variable-group algorithms give the same results as the pair-group ones.
- They always give a uniquely determined solution.
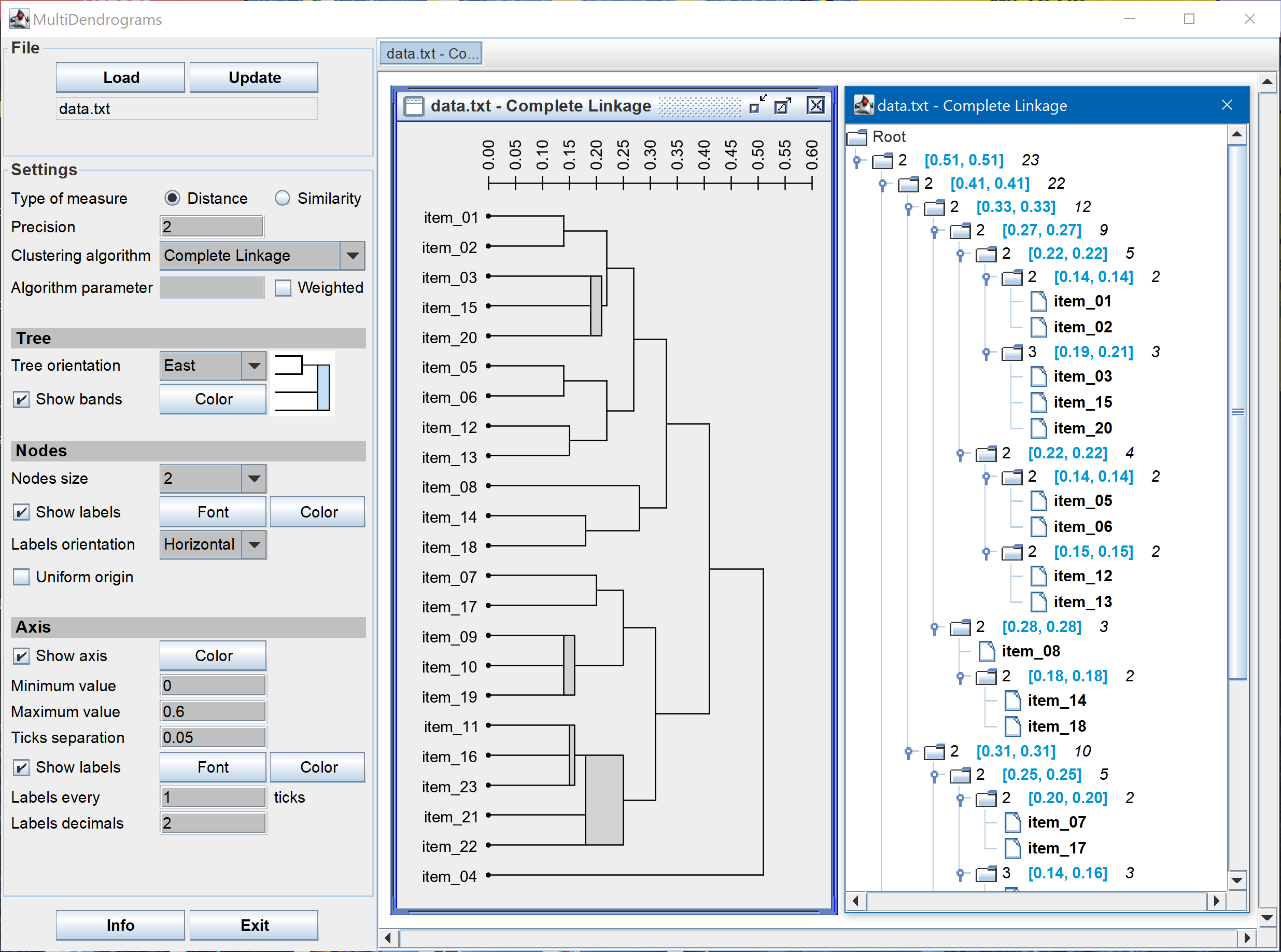
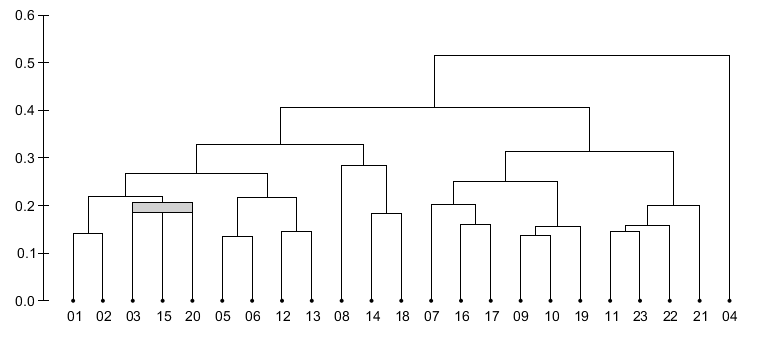
- In the multidendrogram representation for the results one can explicitly observe the occurrence of ties during the agglomerative process. Furthermore, the height of any fusion interval (the bands in the program) indicates the degree of heterogeneity inside the corresponding cluster.
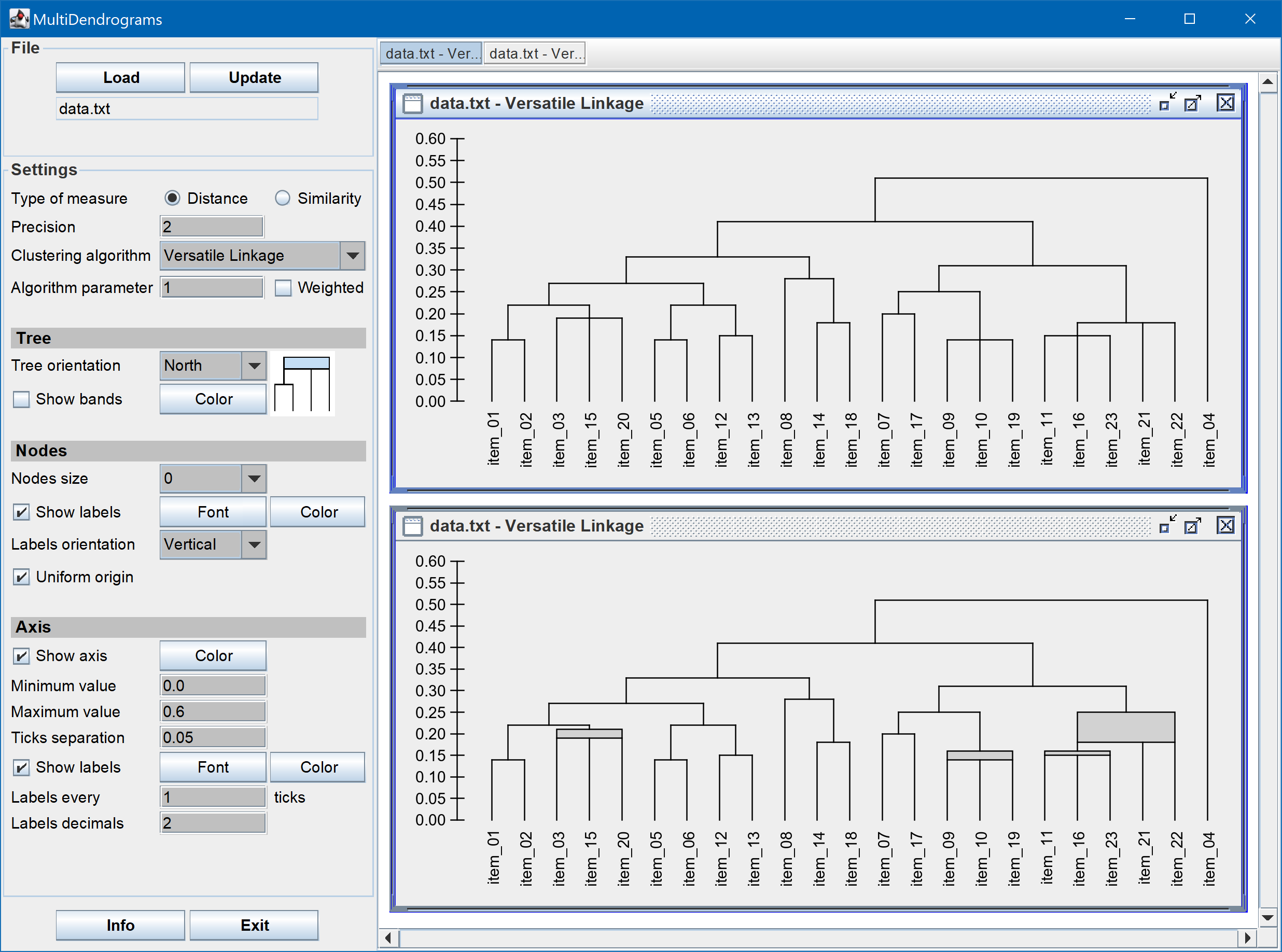
MultiDendrograms also introduces a new parameterized type of hierarchical clustering algorithm called Versatile Linkage [2], which includes Single Linkage, Complete Linkage and Arithmetic Linkage as particular cases, and which naturally defines two new algorithms, Geometric Linkage and Harmonic Linkage (hence the convenience to rename UPGMA as Arithmetic Linkage, to emphasize the existence of different types of averages).
Similar functionality can also be obtained using package mdendro for the R language.
Comparison with other applications
How do other applications deal with ties?
-
Ignore ties, uncommented in their respective manuals:
- Mathematica: Agglomerate and DirectAgglomerate functions in Hierarchical Clustering Package
- MATLAB: linkage function in the Statistics Toolbox
- R: hclust function in the stats package, and agnes function in the cluster package
- Stata: cluster and clustermat commands
-
Report the existence of ties, and break them using the order of the observations in the input file:
- SAS: CLUSTER procedure
-
Break ties using the order of cases in the input file, and recommend the comparison with cases sorted in different random orders:
- SPSS Statistics: Hierarchical Clustering Analysis procedure
How do I know if there are ties in my data?
- Most people would say I do not have problems with tied distances, however you cannot be sure unless the used software explicitly tells you so.
- In MultiDendrograms tied distances can be easily noticed in the dendrogram plots, in the dendrogram navigation window, and in the exported tree files.
How many binary dendrograms may correspond to one MultiDendrogram?
- 6 binary dendrograms for Table 3 in Fatahi et al, Vitis 42 (2003) 185-192.
- 36 binary dendrograms for Supplementary Table 2 in Zdunić et al, Am. J. Enol. Vitic. 64 (2013) 285-290.
- 17900 binary dendrograms for Table 2 in Ibáñez et al, Am. J. Enol. Vitic. 54 (2003) 22-30.
-
760590880 binary dendrograms in Almadanim et al, Vitis 46 (2007) 116-119.
- You may use the Hierarchical_Clustering program in Radatools to count how many binary dendrograms correspond to your data.
References
| [1] |
Solving Non-uniqueness in Agglomerative Hierarchical Clustering Using Multidendrograms
Alberto Fernández and Sergio Gómez Journal of Classification 25 (2008) 43-65 (view) (pdf) (doi) (Springer Nature) |
| [2] |
Versatile linkage: a family of space-conserving strategies for agglomerative hierarchical clustering
Alberto Fernández and Sergio Gómez Journal of Classification 37 (2020) 584-597 (view) (pdf) (doi) (Springer Nature) |
Download
Please cite [1] if you use MultiDendrograms in your publications, and [2] if you use Versatile Linkage:
- Program (manual included): multidendrograms-5.2.1.zip
- Manual: multidendrograms-5.2-manual.pdf
- Source code: multidendrograms-5.2.1-src.zip and at the GitHub repository
Alternatively, you may use mdendro, a package for the R language, or the Hierarchical_Clustering program in Radatools, which is able to calculate MultiDendrograms and also to enumerate or count the corresponding Binary Dendrograms.
Requirements
To run MultiDendrograms it is necessary to have installed a recent version of the Java Runtime Environment (JRE) or the Java Development Kit (JDK). The minimum version of Java needed is Java 8, but it is recommended to use Java 12 or higher. From Java 9 above, the application is scaled using the system scaling, thus avoiding problems with small fonts or windows when using very high resolution (e.g., 4K) screens. You can check if Java is already in your computer and its version following these steps:
- Open a console shell or command prompt (in Windows: Win+R -> cmd -> Enter)
- Type: java -version
We recommend the installation of OpenJDK instead of the Oracle versions of Java, due to the changes in license. There are several options to install OpenJDK 12:
- OpenJDK: requires manual installation (ZIP and TAR.GZ files)
- Zulu: provides installers for the main platforms (MSI for Windows, DMG for MacOS, RPM and DEB for Linux)
- OpenJDKBuilds: installers for Windows
- AdoptOpenJDK: installers for Windows
Installation
No installation needed, just unzip multidendrograms-xxx.zip and run multidendrograms.bat (Windows), multidendrograms.sh (Linux) or multidendrograms.jar (all OS).
Development
You may contribute to the development of MultiDendrograms in GitHub:
- GitHub repository: sergio-gomez / MultiDendrograms
Gallery
History
MultiDendrograms 5.2:
- Input of the power value for Versatile Linkage directly through the algorithm parameter
- Sign of the algorithm parameter unchanged for similarity data
MultiDendrograms 5.1:
- Automatic scaling of graphical user interface
- Upgraded to Java 8; OpenJDK 12 or above recommended
MultiDendrograms 5.0:
- Reorganization of clustering algorithms
- New parameterized Versatile Linkage and Beta Flexible clustering algorithms
- New Geometric Linkage and Harmonic Linkage clustering algorithms
- Calculation of tree balance and space distortion
- Save dendrogram measures to file
MultiDendrograms 4.1:
- Export dendrograms to JSON format
MultiDendrograms 4.0:
- Graphical user interface at different sizes
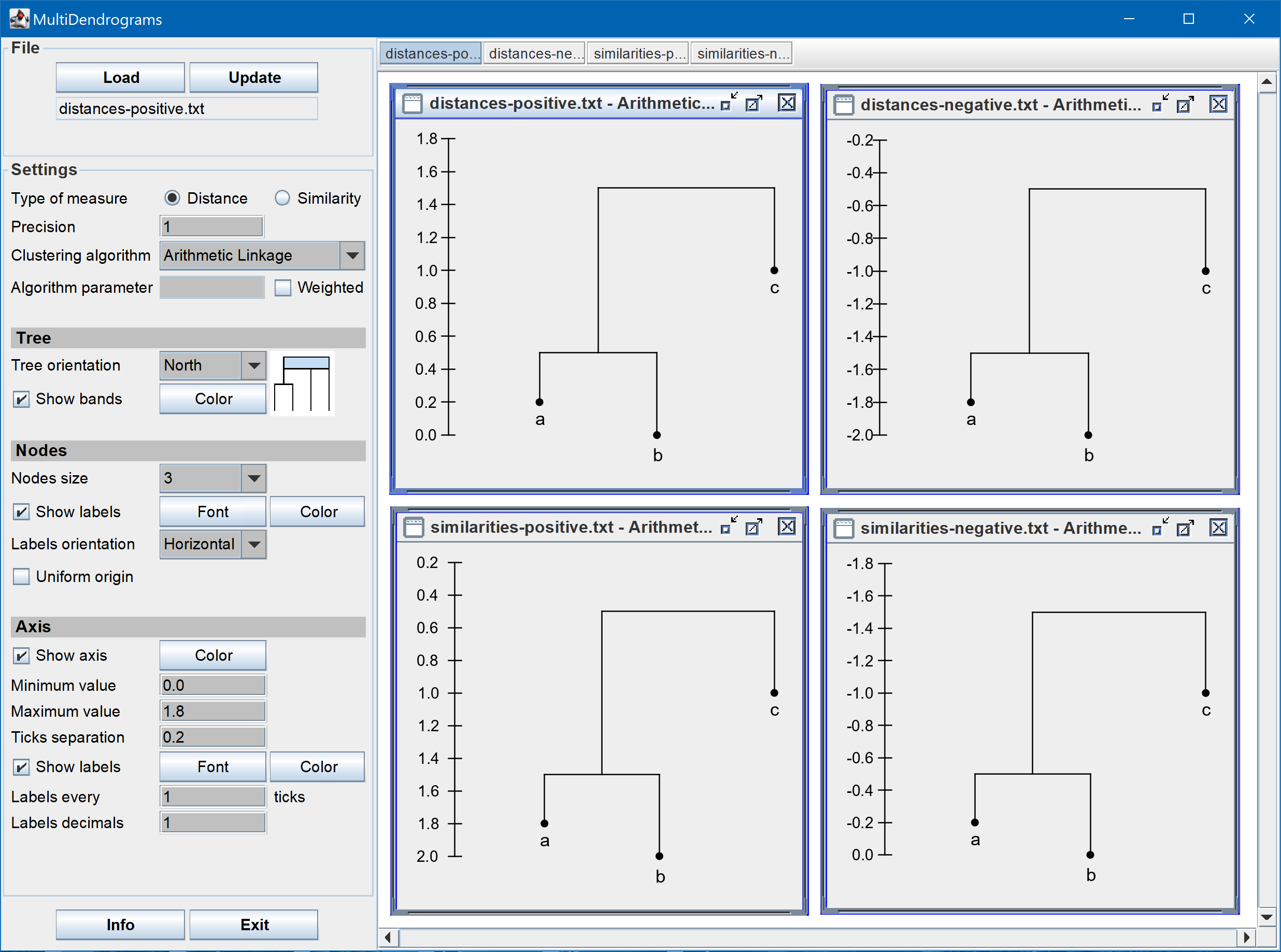
- Positive and negative distances and similarities
- Uniform and non-uniform origin of nodes
- Improved configuration file
- Translation to German
- Improved performance
MultiDendrograms 3.2:
- New format for dendrogram navigation and save as text file
MultiDendrograms 3.1:
- Data in triangular form
MultiDendrograms 3.0:
- Scrollbars in dendrograms panel
- Command-line direct calculation of multidendrogram
- Ward hierarchical clustering
- Check if new version is available
- Confirmation before closing
- Improved performance
- Major source code refactoring
MultiDendrograms 2.1:
- Export dendrograms to Newick format
- Show calculation progress
- Improved GUI
- Improved performance
MultiDendrograms 2.0:
- Completely new multiplatform (Windows, Linux, MacOS, etc.) application
- Added Graphical User Interface (GUI)
- Control of the dendrogram appearance
- Navigation through the dendrogram details
- Accepts distance and similarity matrices
- Export dendrograms to JPG, PNG and EPS
- Calculation of ultrametric deviation measures
MultiDendrograms 1.0:
- Windows command-line application to compute multidendrograms
- Windows command-line application to compute ultrametric matrices
- Windows command-line application to generate EPS plots
Authors
Alberto Fernández:
-
Dept. Enginyeria Química, Universitat Rovira i Virgili, Tarragona, Spain
(email) (ORCID) (Google Scholar) (GitHub)
Sergio Gómez: